JCRB1576 Onda 9
Cell information
Cell type:general cells (View Pricing Information)
| JCRB No. | JCRB1576 | Cell Name | Onda 9 |
|---|---|---|---|
| Profile | Human cell line derived from human glioma. | Other Name | |
| Animal | human | Strain | |
| Genus | Homo | Species | sapiens |
| Sex | M | Age | 58-year-old |
| Identity | available | Tissue for Primary Cancer | |
| Case history | Metastasis | No | |
| Tissue Metastasized | Genetics | P14/P15/P16 deletion, PTEN mutation | |
| Life Span | infinite | Crisis PDL | |
| Morphology | fibroblast-like | Character | - |
| Classify | tumor | Established by | Onda, K. |
| Registered by | Kumanishi, T. | Regulation for Distribution | |
| Comment | Year | 2013 | |
| Medium | DMEM with 10% FBS | Methods for Passages | 0.02% Trypsin + 0.01% EDTA |
| Cell Number on Passage | 1:3 - 1:5 /week | Race | Japanese |
| CO2 Conc. | Tissue Sampling | brain | |
| Tissue Type | anaplastic glioma |
| Detection of virus genome fragment by Real-time PCR | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Detected DNA Virus | tested | Detected RNA Virus | tested | ||||||
| CMV | - | parvoB19 | - | HCV | - | HTLV-1 | - | ||
| EBV | - | HBV | - | HIV-1 | - | HTLV-2 | - | ||
| HHV6 | - | HTLV-1 | - | HIV-2 | - | HAV | - | ||
| HHV7 | - | HTLV-2 | - |
-/negative. +/positive. nt/not tested. (positive (+) does not immediately mean the production of infectious viral particles.) |
|||||
| BKV | - | HIV-1 | - | ||||||
| JCV | - | HIV-2 | - | ||||||
| ADV | - | HPV18 | - | ||||||
| Notes | |||||||||
| Images |
|---|
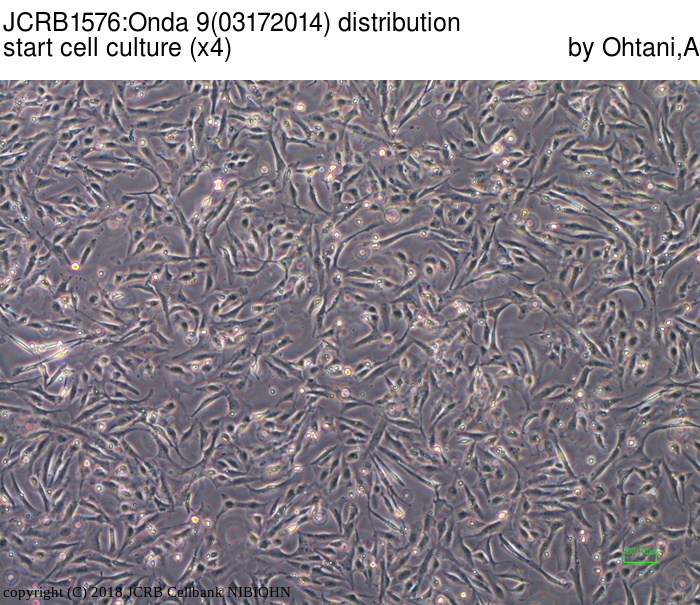 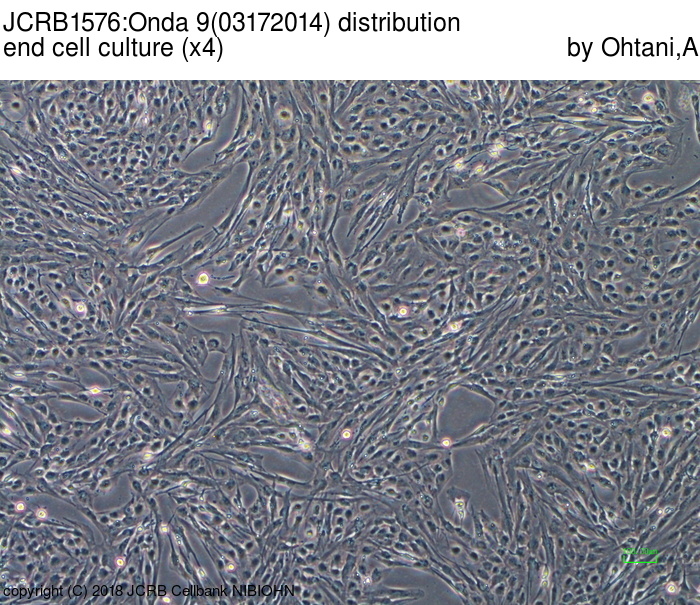 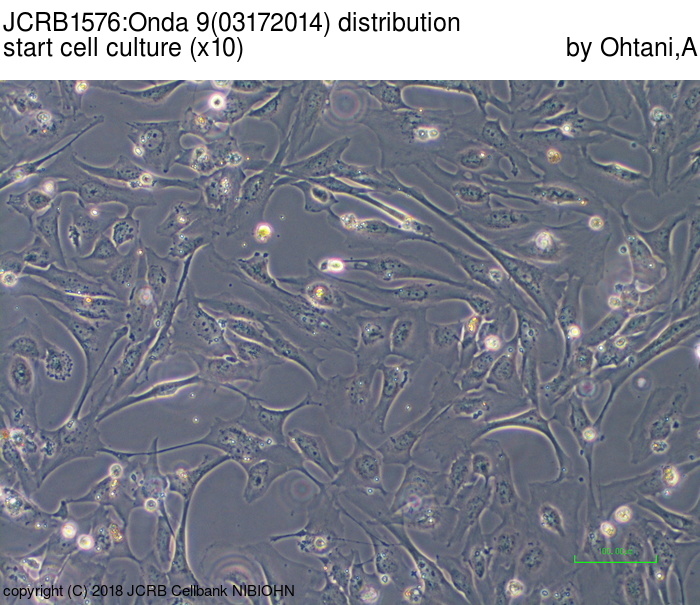 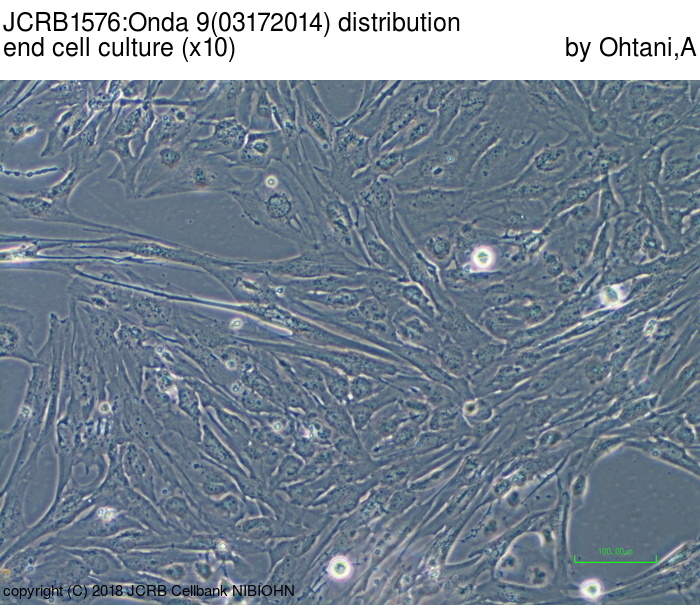 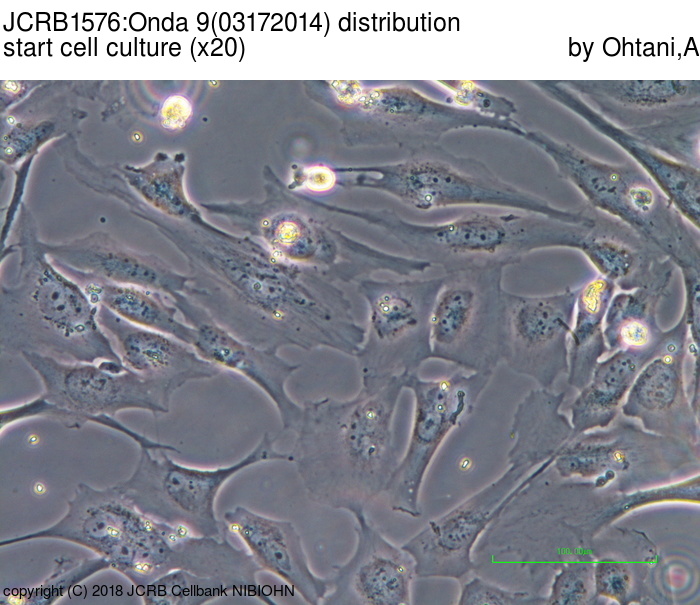 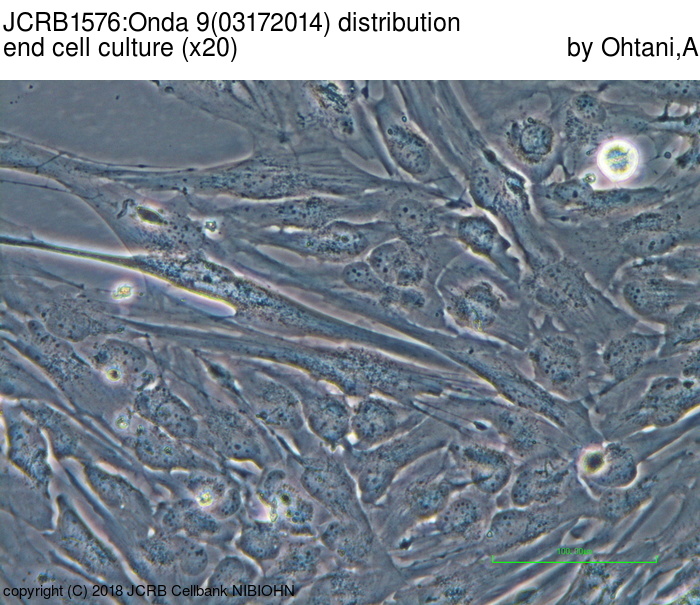 |
LOT Information
Viability/Growth rate/Cell number are represented as actual values measured at lot presentation in JCRB, but are not guaranteed values. Additionally, the doubling time is a rough value measured during passages.| Cell No. | JCRB1576 | Cell Name | Onda 9 |
|---|---|---|---|
| LOT No. | 03172014 | Lot Specification | distribution |
| Medium | DMEM with 10% heat inactivated fetal bovine serum(SIGMA027K03911) | Temperature | 37 C |
| Cell Density at Seeding | 0.99-5.84x10^4cells/sq.cm | Methods for Passages | Cells were harvested after treatment with 0.02%trypsin(GIBCO) and 0.01% EDTA.(Split ratio=1/3-1/5 once a week) |
| Doubling Time | NT | Cell Number in Vial (cells/1ml) | 1.51x10^6 |
| Viability at cell freezing (%) | 97.2 | Antibiotics Used | free |
| Passage Number | p5* | PDL | NT |
| Sterility: MYCOPLASMA | - | Sterility: BACTERIA | - |
| Sterility: FUNGI | - | Isozyme Analysis | NT |
| Chromosome Mode | NT | Chromosome Information | NT |
| Surface Antigen | NT | DNA Profile (STR) | D5S818:9,12 D13S317:11 D7S820:12 D16S539:11,13 VWA:14,18 TH01:9 AM:X,Y TPOX:8,9 CSF1PO:10,12 |
| Adhesion | No | Exoteric Gene | NT |
| Medium for Freezing | Cell Banker BLC-1(Nihon Zenyaku Industries) | CO2 Conc. | 5 % |
| Viability immediately after thawing (%) | Additional information |
| Images |
|---|
      |
| Cell No. | JCRB1576 | Cell Name | Onda 9 |
|---|---|---|---|
| LOT No. | 09172024 | Lot Specification | distribution |
| Medium | Dulbecco's modified Eagle's medium with 10% fetal bovine serum (FBS; Nichirei Cat. # 174112, Lot 20M00K) | Temperature | 37 C |
| Cell Density at Seeding | 0.9 - 1.2 x 10^5 cells/mL | Methods for Passages | Cells were harvested after treatment with 0.02% trypsin and 0.01% EDTA. |
| Doubling Time | approx. 58 hrs. | Cell Number in Vial (cells/1ml) | 1.7 x 10^6 |
| Viability at cell freezing (%) | 65.2 | Antibiotics Used | free |
| Passage Number | Unknown (7 at bank) | PDL | |
| Sterility: MYCOPLASMA | - | Sterility: BACTERIA | - |
| Sterility: FUNGI | - | Isozyme Analysis | |
| Chromosome Mode | Chromosome Information | ||
| Surface Antigen | DNA Profile (STR) | D5S818:9,12 D13S317:11 D7S820:12 D16S539:11,13 VWA:14,18 TH01:9 AM:X,Y TPOX:8,9 CSF1PO:10,12 |
|
| Adhesion | Yes | Exoteric Gene | |
| Medium for Freezing | 10% DMSO, 20% FBS - DMEM | CO2 Conc. | 5% |
| Viability immediately after thawing (%) | 88.0 | Additional information |